Updated: 5th Dec 2019
Key Points
- 2019 WHO report attributes 700,000 deaths annually to antimicrobial-resistance (AMR) organisms and predicts AMR organisms to be the most common cause of human fatalities by 2050.
- Minimum Inhibitory Concentration (MIC)/Breakpoint guidelines differ between the CLSI and EUCAST.
- The aim is to measure the susceptibility of an isolate to a range of antimicrobials and find the MIC.
- Traditional methods are still widely used, there are also many automated antimicrobial susceptibility tests (AST) that return results in faster times.
 Introduction: Resistance to antibiotics can either be naturally occurring for a particular organism or acquired resistance; where misuse of antimicrobials results in a population being exposed to an environment in which organisms that have genes conferring resistance (either spontaneously mutated or through DNA transfer from other resistant cells) have been able to flourish and spread.
Introduction: Resistance to antibiotics can either be naturally occurring for a particular organism or acquired resistance; where misuse of antimicrobials results in a population being exposed to an environment in which organisms that have genes conferring resistance (either spontaneously mutated or through DNA transfer from other resistant cells) have been able to flourish and spread.
What are ESKAPE pathogens? Often referred to in the media as 'superbugs', the ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, Enterobacter spp.) are considered to be the leading cause of hospital-acquired (or nosocomial) infections globally.
Identification of an organism normally goes hand in hand with the AST test, knowing what organism you have isolated together with knowledge of the isolation site, and will give an indication of what type of antibiotics should be considered. The sensitivity of an isolate to a particular antibiotic is measured by establishing the Minimum Inhibitory Concentration (MIC) or breakpoint, this is the lowest concentration (conventionally tested in doubling dilutions) of antibiotic at which an isolate cannot produce visible growth after overnight incubation.
MICs can be determined by agar or broth dilution techniques by following the reference standards established by various authorities such as the Clinical and Laboratory Standards Institute (CLSI, USA), British Society for Antimicrobial Chemotherapy (BSAC, UK), AFFSAPS (France), Deutsches Institut für Normung e.V. (DIN, Germany) & ISC/WHO. These standards are based on pharmacokinetic-pharmacodynamic (PK-PD) properties and mechanisms of resistance. Most European countries follow the MIC cut-offs determined by the European Committee on Antimicrobial Susceptibility Testing (EUCAST). The MIC breakpoints recommended by EUCAST are sometimes higher than CLSI.
MRSA Example: Resistance occurs when the organism has a mecA gene producing an altered penicillin-binding protein, PBP2a (also known as PBP2') and either an oxacillin MIC of 2mg/l or a methicillin MIC of 4mg/l (Always consult the EUCAST and CLSI as the values for susceptibility and resistance change).
Methods in AST and establishing MIC (including rapid automated systems)
Broth Dilution Method - depends upon inoculation at a specific inoculum density of broth media (in tubes or microtitre plates) containing antibiotics at varying levels - usually doubling dilutions are used and after incubation, turbidity is recorded either visually or with an automated reader, and the breakpoint concentration established. Microtitre plates or ready-to-use strips are commercially available with antibiotics ready prepared in the wells. A variation on this approach is the agar dilution method where a small volume of suspension is inoculated onto agar containing a particular concentration of antibiotic when the inoculum has dried the plate is incubated and again examined for zones of growth.
Agar Dilution Method - plates can also be used to determine the MIC, here different concentrations of the antimicrobial are added to non-selective agar before the medium has solidified. The test organism is then prepared and added to the solid agar in different spots, Liofilchem has developed Ready-to-use Agar Dilution Panels, the first and only ready-to-use commercially available kit for performing agar dilution antimicrobial susceptibility testing, in compliance with the CLSI and EUCAST standards.
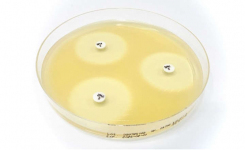
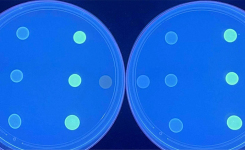
Disk diffusion or the Kirby-Bauer test is one of the classic microbiology techniques and it is still very commonly used. A suspension of the isolate is prepared to a particular McFarland standard, then spread evenly onto an appropriate agar (such as Mueller-Hinton or for a more defined media Iso-Sensitest™ agar) in a petri dish. Disks impregnated with various defined concentrations of different antibiotics are placed onto the surface of the agar. A multichannel disk dispenser can speed up placement of the disks. After incubation, a clear circular zone of no growth in the immediate vicinity of a disk indicates susceptibility to that antimicrobial. Using reference tables the size of the zone can be related to the MIC and results are recorded as to: whether the organism is susceptible (S), intermediately susceptible (I), or resistant (R) to that antibiotic.
There are a number of critical steps in this approach, such as which medium is used; depth and moisture content of the agar in the plate; incubation conditions; accurate inoculum density; disks must be firmly placed in contact with the agar surface otherwise the diffusion rate will not be correct.
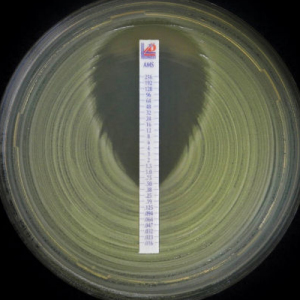
A variation on this approach is to use a strip impregnated along its length, with a gradient of different concentrations of antimicrobial, after incubation this creates an ellipse-shaped zone of no growth, where the ellipse meets the strip, the MIC can be read from the concentration markings on the strip. These are easy to read, no tables need to be referenced to get a MIC value and the test requires fewer manipulations, as one strip will cover the whole concentration range. These again can be manually or instrument read.
Whichever method is used, the result provides a key cutoff point which equates to the minimum inhibitory concentration of antibiotic for that test isolate, and methods initially require a pure culture to be prepared, which may take 1-2 days.
Pre-prepared antibiotic discs with full QC documentation provided by the manufacturer maintain reproducibility and considerably increases assay reliability. Discs should always be manufactured to an acceptable specification e.g. FDA, WHO, DIN. The DIN standard has the tightest range with antibiotic concentrations within 90%-125% of that stated.
Detecting Antibiotic Resistance Markers - Carbapenemases: There are five main carbapenemases (OXA-48-like, KPC, NDM, VIM and IMP) and are produced by carbapenem-resistant Enterobacteriaceae (CRE). The CRE are known to be the most lethal of the ESKAPE pathogens, with a 50% fatality rate by bloodstream infection. There is a number of biotech companies, NG Biotech, Becton and Dickonson (BD) and MAST group that have released handheld carbapenemase diagnostic readers to the market, which give a simple colour change if carbapenemases are present in the sample.
Automated and semi-automated systems perform disk diffusion and broth dilution techniques. Semi-automated comprise of image analysers that will read zones or broth turbidity providing a more objective result and can come with software for automatically interpreting results. For laboratories conducting a large number of assays, there are fully automated systems that are widely used. These typically combine identification with sensitivity testing, and as the whole test is set up and read automatically, not only is the workload reduced but also the result is less subjective and more reproducible. Results are usually faster, with same-day results possible as the instruments monitor growth by taking continuous readings and base results on growth kinetics. Whilst automated systems have many advantages, they can be less flexible in terms of the choice of antibiotics available, consumables costs are usually higher and equipment costs need to be met whether by outright purchase, leasing or reagent rental deals, together with service and maintenance charges.
For both the semi-automated zone readers and the fully automated ID and susceptibility systems, the data collected can be assessed by an expert or smart software systems for interpretation, highlighting unusual anomalous results, suggesting other possible antibiotics to try and can be exported to other LIMS systems for further reporting.