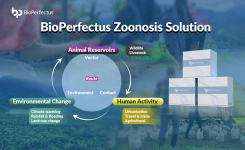
Why this matters:
- Salmonella enterica serovar Dublin is primarily host-adapted to cattle and while it is not one of the top 10 serovars that infect humans, it tends to cause more severe, invasive disease (e.g., bacteremia, septicemia, etc) compared to typical nontyphoidal Salmonella.
- Unlike other serovars, S. Dublin tends to be more homogeneous at the core genome level but exhibits adaptive variation in plasmid content and antimicrobial resistance genes, making it an excellent model for studying microevolution, persistence, and host adaptation.
- The study offers a comprehensive U.S. large-scale genomic snapshot over two decades—something previous studies have often.
- The findings highlight that cattle are likely a key reservoir for AMR S. Dublin, with human and environmental strains being genetically connected.
- Bovine clinical isolates had the highest prevalence of AMR genes and had more frequent carriage of a multidrug-resistance plasmid IncA/C2.
- Bovine strains also showed greater genetic diversity compared to human or environmental strains.
- Despite these differences, a large fraction of strains (72 %) differed from at least one other strain by 20 or fewer SNPs, indicating a high degree of genetic similarity.
- That level of similarity suggests potential for cross-transmission among human, cattle, and environmental reservoirs.
Bigger picture: Salmonella causes approximately 1.35 million infections, 26,500 hospitalizations, and 420 deaths in the United States each year.2 As such, it is one of the leading causes of foodborne illnesses and the leading cause of hospitalizations and deaths linked to foodborne illness.2 This study by Kenney et al.1 underscores the need for integrated surveillance (animals, environment, humans) and rational antibiotic usage policies in livestock to reduce the spillover risk of Salmonella.
References:
- Kenney et al. (2025). “Genomic evolution of Salmonella Dublin in cattle and humans in the United States.” Applied and Environmental Microbiology. Vol. 91, Issue 9: e0068925.
- Center for Disease Control and Prevention (CDC). About Salmonella Infection. Accessed on Oct 9, 2025.
(Image Credit: iStock/SolStock)