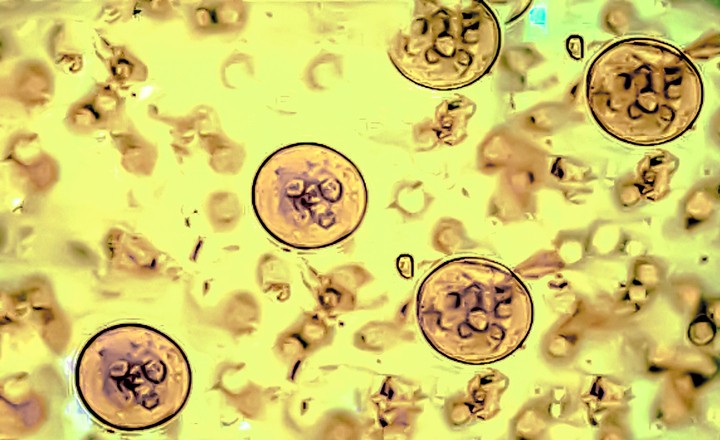
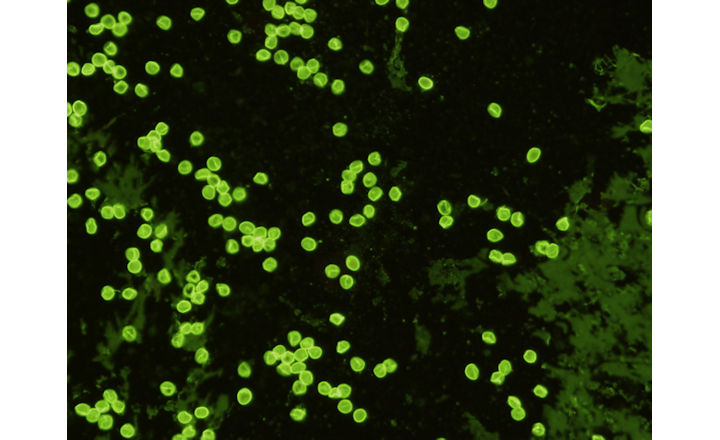
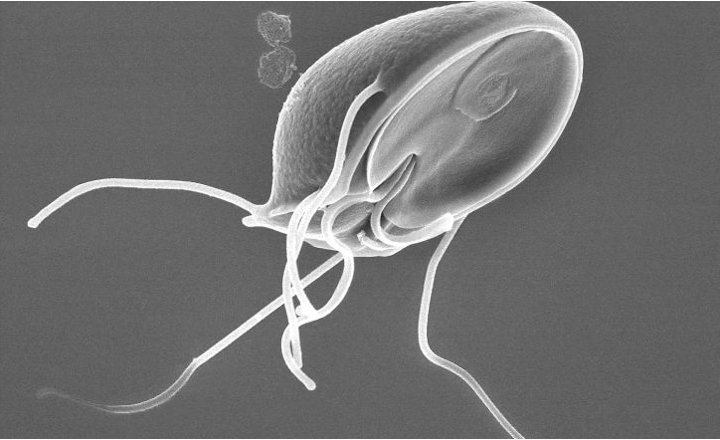
- Cryptosporidium is a major protozoan pathogen associated with foodborne and waterborne outbreaks worldwide, often linked to produce irrigated with contaminated water.
- Its oocysts are highly resistant to environmental stressors and conventional disinfection, complicating control and detection.
- Standard detection methods are labor-intensive and prone to matrix inhibition, limiting surveillance capacity.
- The refined molecular workflow proposed by Schipper et al. enables more consistent detection across agricultural interfaces — a key advance for farm-to-fork risk monitoring.
Key Findings: Schipper et al. (2025) evaluated 11 DNA extractions methods and 3 molecular assays, to optimize a Cryptosporidium detection system which was then assessed by real-time PCR and ddPCR using over 400 environmental (water and soil) and produce (root veggies, fruiting, and leafy greens) samples collected from farms and irrigation systems in South Africa.1
- Improvements in sample concentration, inhibitor removal, and DNA extraction significantly increased oocyst recovery rates compared to conventional methods, including a >10-fold improvement in sensitivity in complex matrices like leafy greens and soil.
- None of the samples tested positive with real-time PCR, while ddPCR detected Cryptosporidium in 13.6% of water, 23.3% of soil, and 34.7% of fresh produce samples.
- Of the water types tested, surface water showed the highest contamination rate (28.6%).
- Of the soil types tested, soil amended with both fertilizer and manure showed the highest contamination rate (45%).
- Among vegetables, roots were most affected (46.7%), followed by fruiting (40%) and leafy greens (30.15%).
- When integrated into existing surveillance frameworks, the method reduced false negatives and allowed consistent detection of low-level contamination.
- These findings also highlight the health risks of Cryptosporidium in food systems.
Bigger Picture: Foodborne Cryptosporidium outbreaks remain largely undetected due to technical challenges in environmental testing. This work provides a validated, field-ready molecular toolkit for integrated monitoring of protozoan pathogens within the water–soil–plant–food nexus. By enabling harmonized detection from source water to edible crops, the approach offers a practical foundation for risk reduction strategies in produce production and irrigation management.
Reference:
- Schipper et al. (2025). Optimized Molecular Detection of Cryptosporidium Within the Water-Soil-Plant-Food Nexus: Advancing Surveillance in Agricultural Systems. Journal of Food Protection. Vol. 88, Issue 9: 100568.