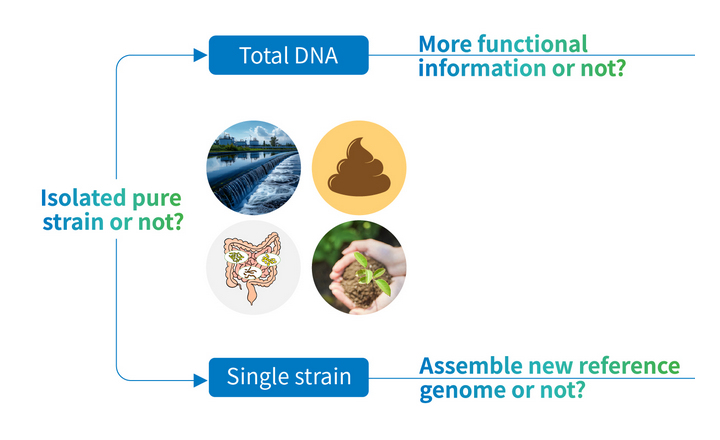
- Whole-genome sequencing (WGS) is increasingly adopted in clinical microbiology for high-resolution pathogen identification, outbreak investigation, virulence profiling, and antimicrobial resistance detection.
- Illumina short-read sequencing has been the clinical standard due to high per-base accuracy but is challenged by assembly of repetitive genomic regions.
- Oxford Nanopore Technologies (ONT) long-read sequencing enables rapid, real-time sequencing with improved assembly continuity, yet historically has had lower raw read accuracy.
- Clinicians and microbiology laboratories need evidence-based guidance on how these platforms perform in bacterial WGS to inform workflow selection for diagnostics, surveillance, and outbreak response.
Key Findings: Purushothaman et al. (2026) conducted a benchmarking study comparing Illumina and ONT sequencing across 20 distinct bacterial genomes representative of clinical pathogen diversity.1 Performance metrics included sequence accuracy, assembly quality, detection of antimicrobial resistance (AMR) determinants, and practical considerations such as time to result and cost.
- Per-base accuracy: Illumina exhibited higher raw per-base accuracy, producing fewer single-nucleotide errors and small indels than ONT in base-level comparisons.
- Assembly completeness: ONT long reads provided enhanced assembly continuity, resolving repetitive regions and structural variants that are challenging with short reads alone.
- Hybrid assembly advantage: Combining Illumina and ONT data (hybrid assemblies) yielded the most complete and accurate genomes, outperforming either technology alone for structural resolution and AMR gene context.
- Antimicrobial resistance detection: Both platforms reliably detected known AMR determinants; Illumina’s higher fidelity facilitated confident single-nucleotide polymorphism (SNP) calls, while ONT’s long reads improved linkage of resistance genes within mobile genetic elements.
- Time and operational considerations: ONT’s rapid sequencing and real-time analysis shortened turnaround time, advantageous for urgent clinical contexts, whereas Illumina’s batching and longer workflows may suit high-throughput reference labs.
Bigger Picture: This benchmarking study provides actionable insights for clinical microbiology laboratories integrating whole-genome sequencing. Illumina remains a gold standard for high-accuracy base calling and mature informatics pipelines. ONT augments WGS with rapid long reads and improved assembly of complex genomic regions. Hybrid sequencing strategies combine the strengths of both to deliver the most robust genomes for diagnostics, epidemiology, and AMR characterization. As sequencing costs decline and bioinformatics tools mature, choosing appropriate platforms or combinations will depend on the application — urgent outbreak detection, high-resolution phylogenomics, or comprehensive AMR profiling — and the laboratory’s operational priorities.
(Image Credit: iStock/JuSun)
References:
- Purushothaman et al. 2026. Benchmarking Illumina and Oxford Nanopore Technologies (ONT) Sequencing Platforms for Whole Genome Sequencing of Bacterial Genomes and Use in Clinical Microbiology. BMC Medical Genomics.