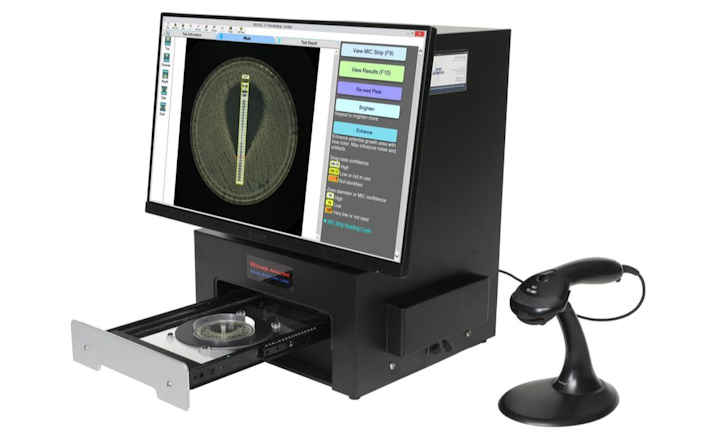
Arc Bio have launched proprietary antimicrobial resistance software, marking the first in what will be a series of new next-generation sequencing (NGS)-based products aimed at revolutionizing the field of pathogen detection. Galileo™ AMR is an antimicrobial resistance (AMR) detection software that provides fast, accurate annotations for any gram-negative bacterial DNA sequence in less than five minutes.
"At Arc Bio, we are on a mission to improve the human condition by delivering new tools that profoundly change how disease is diagnosed, treated and managed. The launch of Galileo™ AMR is our first step towards achieving this goal," said Arc Bio Chief Executive Officer Dr. Todd Dickinson, a founding scientist of Illumina. "As the CDC reports, every year over 2 million people are infected by antibiotic resistant bacteria, causing more than 23,000 deaths in the U.S.1 Rapid identification of various strains of antimicrobial resistance, and better understanding their transmission and evolution, is vital to protecting public health."
Arc Bio, an EdenRoc Sciences Company, was co-founded by Dr. Carlos Bustamante, a population geneticist, Professor of Biomedical Data Science, Genetics, and (by courtesy) Biology at Stanford University, where he serves as the Inaugural Chair of the Department of Biomedical Data Science; and Dr. David Andrew Sinclair, a Professor of Genetics at Harvard University and founding Director of the Paul F. Glenn Laboratories for the Biological Mechanisms of Aging. Both serve on Arc Bio's Scientific Advisory Board.
"Our goal at Arc Bio is to revolutionize pathogen detection by developing a unique NGS lab workflow and software solution that allows for smarter and simple to use analysis," Bustamante said. "Our current emphasis is on assisting those in public health and life science research who study antimicrobial resistance transmission and evolution of gram-negative bacteria."
"We know that there are more efficient ways to detect and annotate resistance in order to better protect and improve public health," Sinclair said. "As Arc Bio evolves, we aim to play an increasingly significant role in combatting the global challenges of infectious disease and antibiotic resistance."
Formerly known as MARA and acquired from Spokade – a leading company in the field of monitoring and controlling antimicrobial resistance -- Galileo™ AMR features the most extensive archive of expert-validated gram-negative AMR genes, cassettes and other mobile elements.2,3
The software incorporates advanced analytics in a user-friendly interface to:
- quickly and efficiently detect AMR in gram-negative bacteria,
- precisely annotate AMR genes and mobile elements in DNA sequences of any length, and
- provide accurate annotation of plasmid AMR insert sequences.
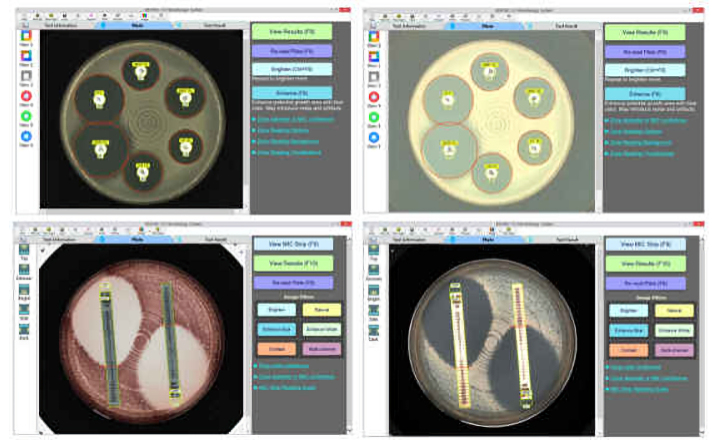
The Galileo™ AMR cloud-based proprietary software provides precise AMR annotations, quick and reliable results, an intuitive user interface that does not require bioinformatics expertise, and detailed, easy-to-understand diagrams.
References:
- 1. Centers for Disease Control and Prevention https://www.cdc.gov/drugresistance/index.html.
- 2. Tsafnat G, Copty J, Partridge SR. RAC: Repository of Antibiotic resistance Cassettes. Database. 2011; bar054. doi:10.1093/database/bar054/470201
- 3. Partridge SR, Tsafnat G. Automated annotation of mobile antibiotic resistance in Gram-negative bacteria: the Multiple Antibiotic Resistance Annotator (MARA) and database. 2018 Apr 1;73(4):883-890. doi: 10.1093/jac/dkx51