Comparison between Droplet Digital PCR (ddPCR) and qPCR in screening and confirming Shiga Toxin-Producing E. coli (STEC) with linked and unlinked stx and eae in Beef Matrices
Current qPCR-based methods for screening STEC have the common challenge of differentiating between samples where a single organism contains both stx and eae (true positive, linked virulence) from samples in which stx and eae reside in different organisms (false positive, unlinked virulence). Bio-Rad offers a Droplet Digital PCR solution, the dd-Check STEC Solution, that demonstrates the capacity in virulence linkage analysis by partitioning intact cells into droplets where cell lysis and PCR amplification occur, enhancing the test accuracy by reducing false-positive reactions. To evaluate the ability of ddPCR technology to detect and distinguish E. coli cells with linked and unlinked virulence compared to qPCR technology, a collaborative study between Food Safety Net Services and Bio-Rad was performed.
A set of three matrices, beef trim, ground beef and MicroTally (n=30), were inoculated at < 5 CFU per 375 g sample. For each sample type, 15 samples were inoculated with one regulated STEC strain with linked virulence genes (stx and eae in the same cell), and 15 samples were inoculated with a cocktail of two regulated serotypes with unlinked virulence genes (one with stx only and one with eae only).
All samples were processed following two timelines for evaluation of the ddPCR technology as a primary screening assay compared to qPCR and a confirmatory assay, illustrated in Fig. 1. Following the timeline for using ddPCR as primary screening, samples were processed for detection of stx and eae via qPCR (iQ-Check STEC VirX Kit, Bio-Rad) and ddPCR technologies (dd-Check STEC Solution, Bio-Rad) after a 16 hr enrichment. Following the timeline for using ddPCR as a confirmatory assay, samples were processed for primary screening using qPCR after an 8 hr enrichment, followed by confirmation via ddPCR with or without regrowth according to the criteria demonstrated in Fig 1.
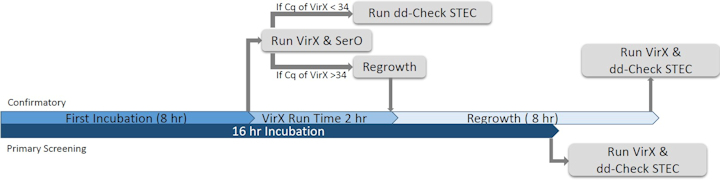
Figure 1
Cultural confirmation of STEC in all samples was performed following the USDA MLG 5C.00 protocol. The results obtained by qPCR and ddPCR assays were compared against the reference method for accuracy in screening and distinguishing STEC with linked and unlinked virulence genes.
Results demonstrated that the ddPCR technology was able to individually identify the presence of stx and eae and verify the co-existence of these two virulence genes via linkage analysis (Fig 2).
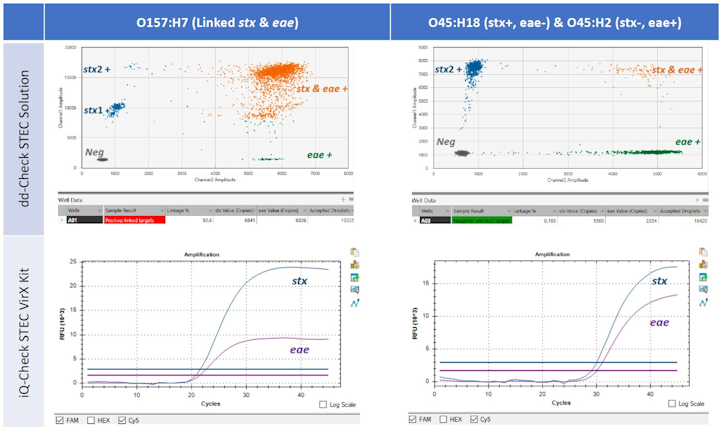
Figure 2
The ability of ddPCR technology as a primary screening assay to identify the co-existence of stx and eae in a single bacterium enhances the accuracy of STEC testing by reducing number of false positive reactions. In this study, ddPCR technology also proved useful for culture-independent confirmation following primary screening by qPCR assays.
For more information visit: bio-rad.com/ddcheck or click the Request Information button below.