Sartorius Stedim Bio...
Smart Tools for Micr...
20th September 2017 Content supplied by: Sartorius
Benchmarking of the Microsart® AMP Mycoplasma Kit
Introduction Among the world’s smallest bacteria, Mycoplasmas are capable of independent reproduction. They belong to the Mollicutes class and exhibit very slow, parasitic growth. The contamination of cell cultures remains a major problem. An array of physiological and biochemical parameters are affected by the presence of Mycoplasma in cell cultures. The infection causes changes in metabolism, growth, viability, macromolecule synthesis, morphology etc. and therefore a sensitive routine testing for contamination of cell cultures is essential.
The traditional growth-based method requires a cultivation time of at least 28 days before contamination can be ruled out with certainty. In comparison, Nucleic Acid Amplification Techniques (NAT) allow a reduction in time to result to just hours. To use as an alternative to the culture method, any NAT test system must be shown to detect 10 CFU/mL of Mycoplasma.
For this reason, the ability of three different Mycoplasma PCR kits to detect samples spiked with not higher than 10 CFU per ml in Dulbecco’s Modified Eagle Medium was tested in this study.
Experimental Set-up and Results During this study the Microsart® AMP Mycoplasma kit inclusive sample preparation with the kit Microsart® AMP Extraction (Sartorius Stedim Biotech GmbH) were benchmarked with two other Mycoplasma detection kits which are also based on DNA isolation with subsequent Real-time PCR analysis. As test material the 10 CFU Sensitivity Standards Mycoplasma arginini (NCTC code 10129), Mycoplasma orale (NCTC code 10112), Spiroplasma citri (NCTC code 10164), (Sartorius Stedim Biotech GmbH) were used.
These lyophilized standards can be easily rehydrated by adding 1 ml of sample matrix to achieve a concentration of 10 CFU/ml. These preparations are intended for safe and reliable validation of Mycoplasma PCR assays. In these comparative studies DMEM with 5 % FCS was used as sample matrix.
For all three assays, the DNA isolation process and the subsequent PCR set-up was done according to the manual of the manufacturer. The Microsart® AMP Extraction kit is based on a convenient silica column based protocol which can be completed within 30 minutes. The DNA isolation procedures of the competing products are based on DNA precipitation on a Magnetic Bead based protocol. Two of the three evaluated PCR assays include highly specific TaqMan probes; one of them is the Microsart® AMP Mycoplasma.
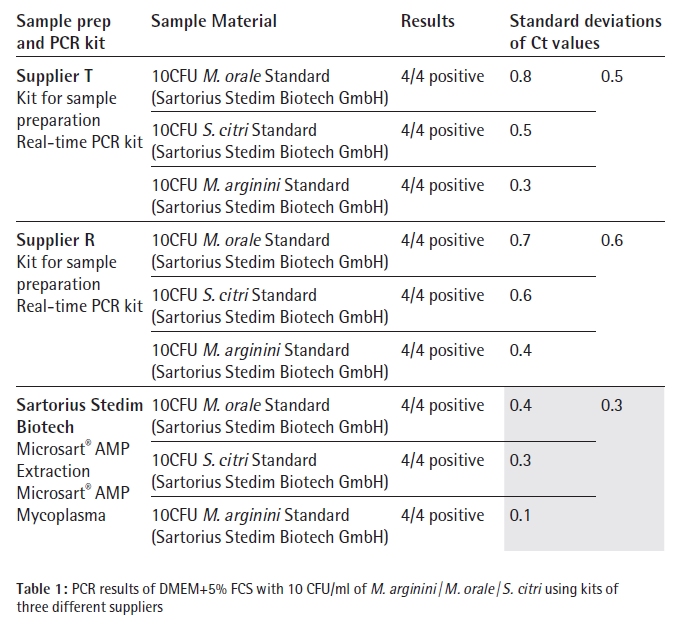
The third assay includes SYBR Green in the detection mix which makes a melting curve analysis necessary after the DNA amplification cycles to differentiate between unspecific amplified DNA and Mycoplasma amplicon DNA. Each species, M. arginini, M. orale and S. citri, was tested four times with each kit, which included the whole process of DNA isolation, polymerase chain reaction and data analysis. The results are listed in Table 1.
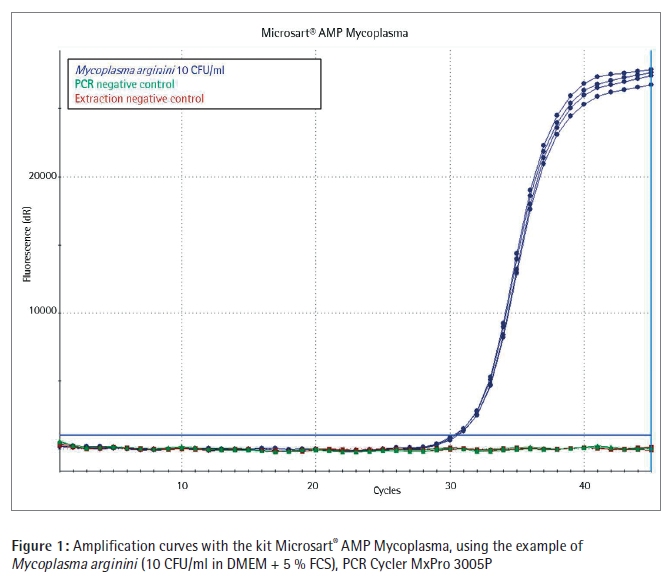
Conclusion In these studies Sartorius Microsart® AMP Extraction and Microsart® AMP Mycoplasma were benchmarked with Mycoplasma Real-time PCR kits incl. sample preparation of two other suppliers.
Each of the 36 DNA extracts showed positive signals in the subsequent PCR analysis. So all tested products are able to detect Mycoplasma contaminations with a sensitivity of at least 10 CFU per ml. Having a closer look at the standard deviation for the results of one supplier, the Microsart® AMP Extraction followed by amplification with the Microsart® AMP Mycoplasma kit has led to the least variability and highest reproducibility. The PCR kits, but predominately the three different sample preparation methods might have contributed to the difference in reproducibility.
Silica-membrane based procedures are known to be very robust and reliable even when processing highly complex sample matrices. DNA precipitation or Magnetic bead based protocols tend to be more laborious and less robust. Another fact which also contributes to the reliability of the Microsart® Mycoplasma product range is that all PCR reagents can be stored at 4 to 8 °C. So, a short interruption in the cool chain will not lead to undesired thawing of the reagents which can have negative effects on the sensitivity of the PCR assay.
In contrast the Mycoplasma detection kits of supplier T and supplier R have to be stored frozen at – 20°C. When planning tests with these products additional waiting time for thawing of the PCR reagents has to be scheduled in. Furthermore, it has to be considered that if a kit which needs to be stored at – 20°C was not used up completely within one PCR run, then several thawing and freezing cycles will also have negative effects on the PCR results.
Tags:
Date Published: 20th September 2017
Source article link: View
Sartorius Stedim Biotech Opens New
Smart Tools for Microbiological Quality